Distribution of antibiotic-resistant strains of Klebsiella pneumoniae in modern conditions
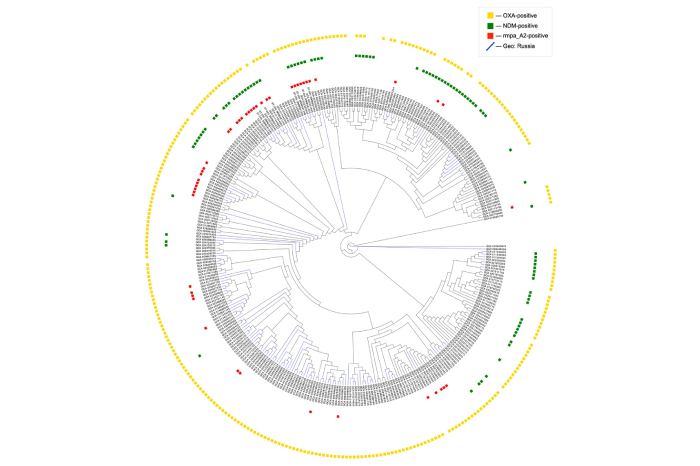
An analysis of 46,000 Klebsiella pneumoniae genomes revealed trends in the spread of antibiotic resistance genes and the emergence of strains that combine multidrug resistance with a hypervirulent genotype.
Klebsiella pneumoniae is a common opportunistic pathogen that can cause severe nosocomial infections in healthy individuals.
Specialists from the Laboratory for Innovative Methods of Microbiological Monitoring of the WCRC for Personalised Medicine, together with colleagues from the Mechnikov State Medical University, conducted an analysis of all Klebsiella pneumoniae genome sequences available in NCBI GenBank. The aim of the study was to perform genetic typing and search for antibiotic resistance genes and virulence determinants in Klebsiella strains isolated from different regions of the world over the last 20 years.
The analysis showed a rapid increase in the proportion of strains carrying the most important resistance genes to the last-line antibiotics - carbapenems. In particular, the frequency of detection of new Delhi metallo-β-lactamase (NDM) genes was considered.
The most pronounced growth rate of strains carrying NDM is observed during the COVID-19 pandemic: from 17% in 2020 to 30% in 2021 and 38% in 2022. During the same period, there is a pronounced increase in the number of hypervirulent Klebsiella strains with multidrug resistance, including among strains circulating in Russia.
As a result of the analysis, it was also possible to determine the geographical characteristics of the distribution of the main genetic lineages of the pathogen. Sequence type 395 was characteristic of Russia, accounting for one third of all sequence types. At the same time, Klebsiella of this genetic line are much less common in other regions of the world.
Analysis of the genomes of epidemiologically
significant healthcare-associated pathogens is helping to identify patterns in
the spread of antibiotic resistance and improve approaches to contain it.
